Polymerase chain reaction (PCR) is a method for rapidly making millions to billions of copies of a DNA sample. This method allows scientists to amplify a small piece of DNA or even a part of it. PCR was founded in 1983 by Kary Mullis (who was a biochemist) and his colleagues. Mullis and Michael Smith (who developed other methods of DNA manipulation) won the Nobel Prize in Chemistry in 1993 for their work. Today, this technique has wide applications in research centers, medical laboratories, analysis of ancient DNA fragments and in forensic medicine.
PCR technique has 2 main reagents:
- Primer: It is a short deoxyribonucleotide fragment, which is also called an oligonucleotide, and is complementary to a part of the desired DNA.
- Heat-resistant DNA Polymerase enzyme
Applications of PCR:
Applications of the technique include DNA cloning for sequencing, gene cloning and manipulation, gene mutagenesis; construction of DNA-based phylogenies, or functional analysis of genes; diagnosis and monitoring of genetic disorders; amplification of ancient DNA;[5] analysis of genetic fingerprints for DNA profiling (for example, in forensic science and parentage testing); and detection of pathogens in nucleic acid tests for the diagnosis of infectious diseases.
A basic PCR set-up requires several components and reagents, including:
- a DNA template that contains the DNA target region to amplify
- a DNA polymerase; an enzyme that polymerizes new DNA strands; heat-resistant Taq polymerase is especially common, as it is more likely to remain intact during the high-temperature DNA denaturation process
- two DNA primers that are complementary to the 3' (three prime) ends of each of the sense and anti-sense strands of the DNA target (DNA polymerase can only bind to and elongate from a double-stranded region of DNA; without primers, there is no double-stranded initiation site at which the polymerase can bind); specific primers that are complementary to the DNA target region are selected beforehand, and are often custom-made in a laboratory or purchased from commercial biochemical suppliers
- deoxynucleoside triphosphates, or dNTPs (sometimes called "deoxynucleotide triphosphates"; nucleotides containing triphosphate groups), the building blocks from which the DNA polymerase synthesizes a new DNA strand
- a buffer solution providing a suitable chemical environment for optimum activity and stability of the DNA polymerase
- bivalent cations, typically magnesium (Mg) or manganese (Mn) ions; Mg2+ is the most common, but Mn2+ can be used for PCR-mediated DNA mutagenesis, as a higher Mn2+ concentration increases the error rate during DNA synthesis;[11] and monovalent cations, typically potassium (K) ions
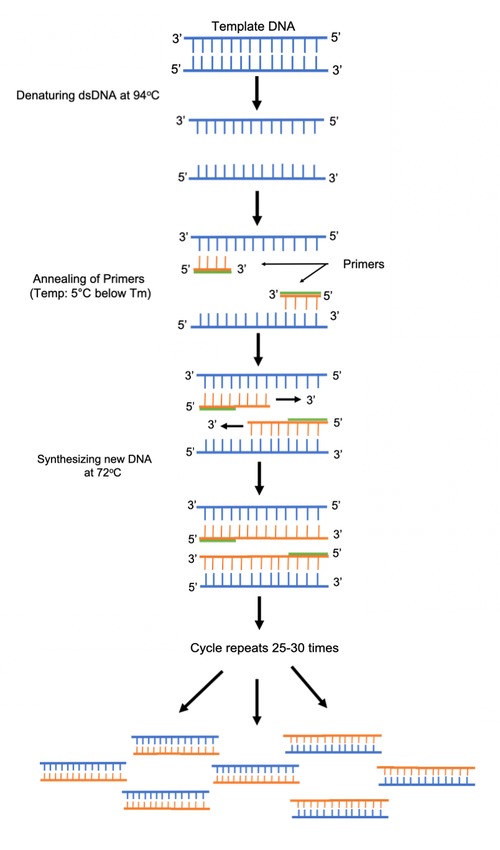
The individual steps common to most PCR methods are as follows:
- Initialization: This step is only required for DNA polymerases that require heat activation by hot-start PCR. It consists of heating the reaction chamber to a temperature of 94–96 °C (201–205 °F), or 98 °C (208 °F) if extremely thermostable polymerases are used, which is then held for 1–10 minutes.
- Denaturation: This step is the first regular cycling event and consists of heating the reaction chamber to 94–98 °C (201–208 °F) for 20–30 seconds. This causes DNA melting, or denaturation, of the double-stranded DNA template by breaking the hydrogen bonds between complementary bases, yielding two single-stranded DNA molecules.
- Annealing: In the next step, the reaction temperature is lowered to 50–65 °C (122–149 °F) for 20–40 seconds, allowing annealing of the primers to each of the single-stranded DNA templates. Two different primers are typically included in the reaction mixture: one for each of the two single-stranded complements containing the target region. The primers are single-stranded sequences themselves, but are much shorter than the length of the target region, complementing only very short sequences at the 3' end of each strand.
It is critical to determine a proper temperature for the annealing step because efficiency and specificity are strongly affected by the annealing temperature. This temperature must be low enough to allow for hybridization of the primer to the strand, but high enough for the hybridization to be specific, i.e., the primer should bind only to a perfectly complementary part of the strand, and nowhere else. If the temperature is too low, the primer may bind imperfectly. If it is too high, the primer may not bind at all. A typical annealing temperature is about 3–5 °C below the Tm of the primers used. Stable hydrogen bonds between complementary bases are formed only when the primer sequence very closely matches the template sequence. During this step, the polymerase binds to the primer-template hybrid and begins DNA formation.
- Extension/elongation: The temperature at this step depends on the DNA polymerase used; the optimum activity temperature for the thermostable DNA polymerase of Taq polymerase is approximately 75–80 °C (167–176 °F), though a temperature of 72 °C (162 °F) is commonly used with this enzyme. In this step, the DNA polymerase synthesizes a new DNA strand complementary to the DNA template strand by adding free dNTPs from the reaction mixture that is complementary to the template in the 5'-to-3' direction, condensing the 5'-phosphate group of the dNTPs with the 3'-hydroxy group at the end of the nascent (elongating) DNA strand. The precise time required for elongation depends both on the DNA polymerase used and on the length of the DNA target region to amplify. As a rule of thumb, at their optimal temperature, most DNA polymerases polymerize a thousand bases per minute. Under optimal conditions (i.e., if there are no limitations due to limiting substrates or reagents), at each extension/elongation step, the number of DNA target sequences is doubled. With each successive cycle, the original template strands plus all newly generated strands become template strands for the next round of elongation, leading to exponential (geometric) amplification of the specific DNA target region.
The processes of denaturation, annealing and elongation constitute a single cycle. Multiple cycles are required to amplify the DNA target to millions of copies. The formula used to calculate the number of DNA copies formed after a given number of cycles is 2n, where n is the number of cycles. Thus, a reaction set for 30 cycles results in 230, or 1,073,741,824, copies of the original double-stranded DNA target region.
- Final elongation: This single step is optional, but is performed at a temperature of 70–74 °C (158–165 °F) (the temperature range required for optimal activity of most polymerases used in PCR) for 5–15 minutes after the last PCR cycle to ensure that any remaining single-stranded DNA is fully elongated.
- Final hold: The final step cools the reaction chamber to 4–15 °C (39–59 °F) for an indefinite time, and may be employed for short-term storage of the PCR products.
There are two methods for measuring amplified DNA for Real time PCR:
The first method: using non-specific fluorescent markers using a dye like (SYBR® Green)
In this method, the observation of any double-stranded DNA sequence is provided. It does not need a probe and thus reduces the measurement setup and running costs. Also, multiple dyes can be attached to a single amplified molecule, which increases the sensitivity for detecting amplified products.
In the real-time SYBR Green PCR reaction, a template that contains the desired target sequence, as well as primers or small pieces of DNA for amplification, four bases that form DNA or dNTPs (dTTP, dATP, dCTP, dGTP) and the desired DNA polymerase enzyme are needed SYBR Green dye is usually included in the reaction mixture containing DNA polymerase enzyme.
Real time polymerase chain reaction includes a series of repeated cycles (usually 35-40 cycles) and each of these cycles includes the four steps described above.
In this type of method, following elongation, SYBR Greens bind to all newly synthesized double-stranded DNA complexes and emit fluorescence light. As the PCR cycle continues, fluorescent light is accumulated and measured at the end of each PCR cycle.
The second method: using specific fluorescent markers using a probe
Real-time PCR based on the probe provides the possibility of hybridization. The targeted nature of probe-based Real-Time PCR results in low background and eliminates the presence of false positives. You can also label the probes with different, detectable reporter dyes to allow amplification of two separate sequences in one reaction tube.
Today, various companies around the world produce and sell PCR machines and their accessories. These devices have different features and levels of accuracy. China Tianlong Company is a manufacturer of molecular equipment, including PCR thermocycler and complete PCR rail and their accessories.
These devices can be used in research and medical diagnosis centers. Vesta Tejhiz Part Company is the exclusive sales and after-sales service representative of this company. By employing experienced experts, this company has always tried to gain the satisfaction of respected experts and researchers.













